Bayesian Spatial-Temporal Modeling & Methodological Advancements in Public Health Research
This research area focuses on advanced spatial-temporal analysis and methodological innovations to address various public health challenges. The research primarily focuses on understanding dynamic trends, patterns, and disparities in health-related issues through a spatial-temporal lens. These methodological contributions offer valuable insights and robust frameworks, aiding researchers, practitioners, and policymakers in devising effective and equitable health strategies and interventions based on a nuanced understanding and analysis of public health data.
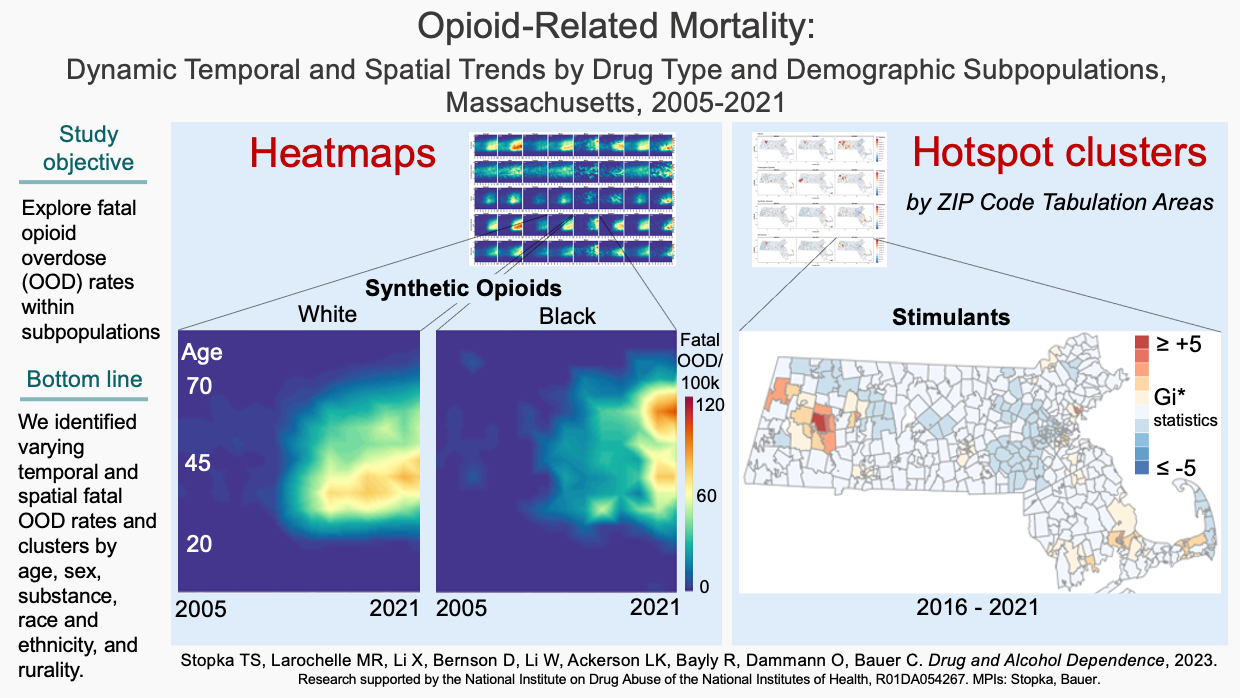
Use novel visulization and analysis, we uncovered new time varying and spatial patterns in fatal OOD rates not previously reported. Identified shifts in fatal OOD rates by sex, age, and race/ethnicity can inform location-specific field actions targeting subpopulations at disproportionally high risk.
(Check our publication:
Opioid-related mortality: Dynamic temporal and spatial trends by drug type and demographic subpopulations, Massachusetts, 2005-2021. )
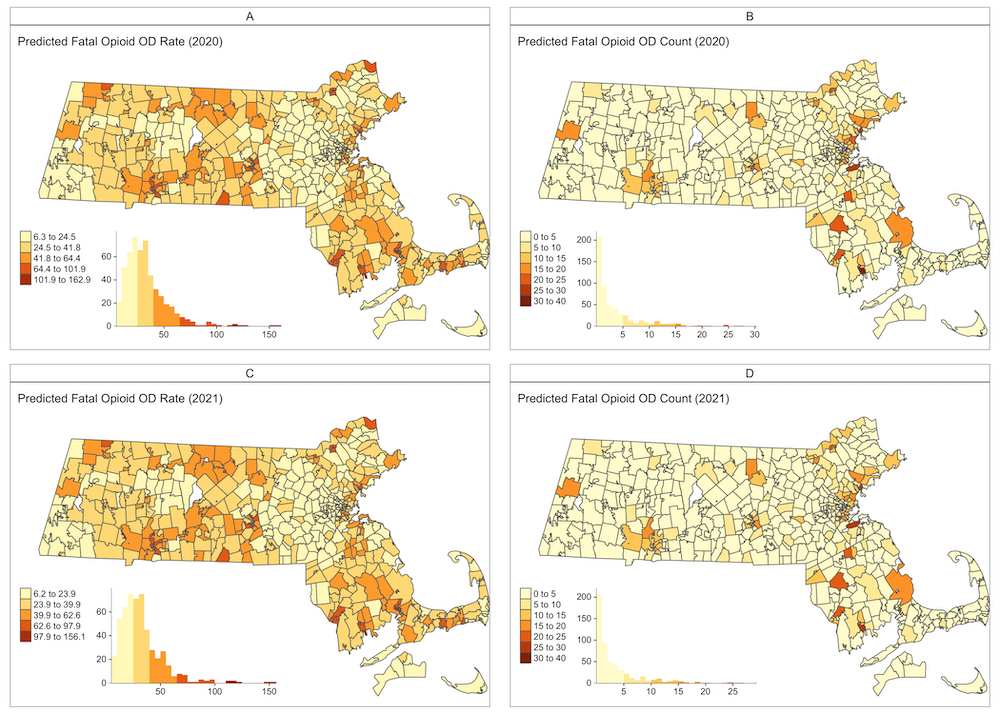
We have highlighted how Bayesian spatiotemporal models have been successfully used to predict opioid-related deaths,
at the ZIP Code level in Massachusetts. The aim is to provide information for public health interventions.
By analyzing data from 2005 to 2019 and incorporating demographic and socioeconomic factors, our models have shown improved
accuracy especially when considering the spatial dynamics of neighborhoods. Interestingly our models performed better in urban areas
compared to rural regions with lower population density. Predictions indicated a potential stabilization in opioid-related deaths
for 2020-2021 based on past trends. These models can guide public health decisions and can be replicated in other jurisdictions
and at varying temporal and geographical levels, emphasizing the need for collaborative modeling efforts in opioid overdose predictions.
(Check our publication:
Small area forecasting of opioid-related mortality: Bayesian spatiotemporal dynamic modeling approach. )
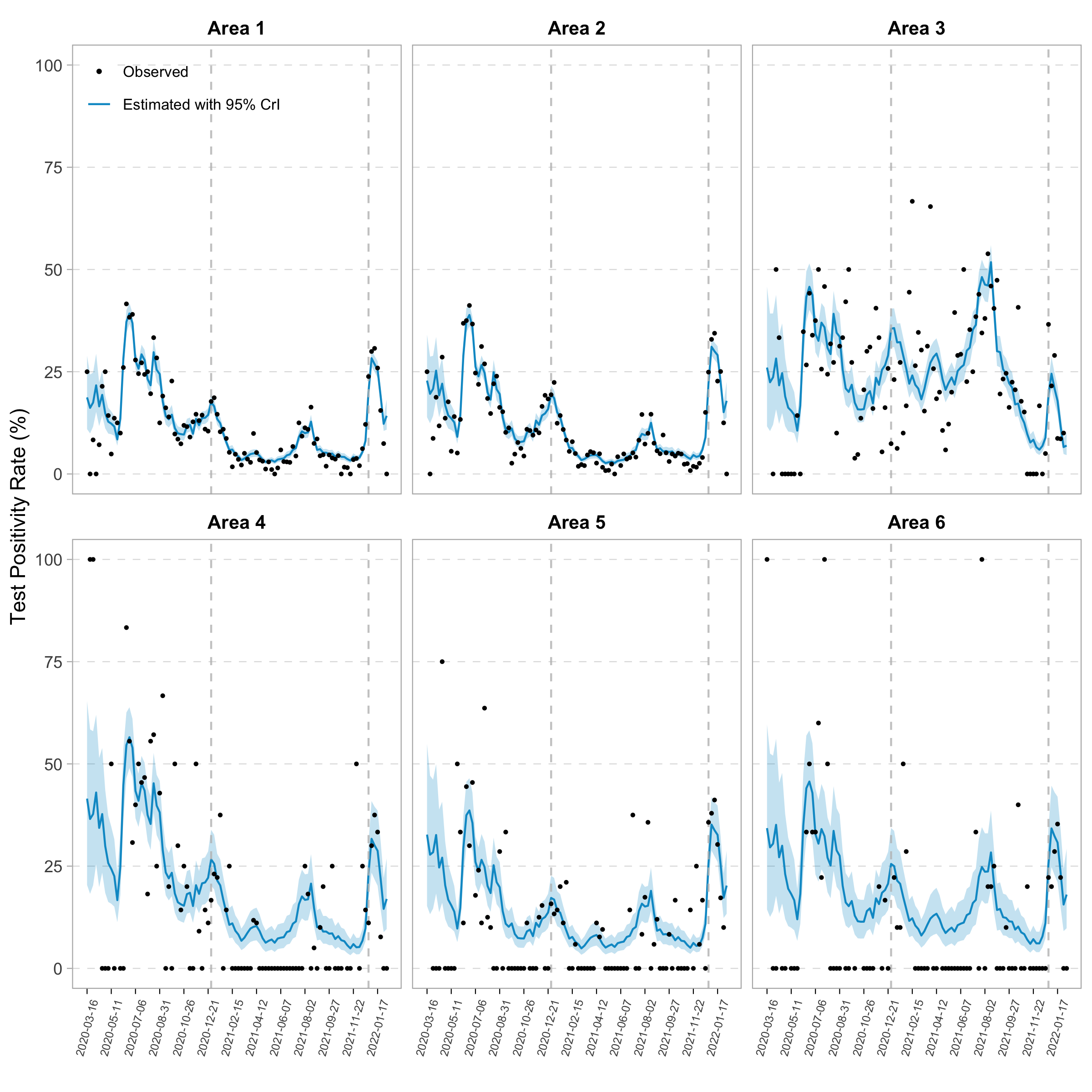
We proposed a novel Bayesian spatial-temporal approach to identify and quantify severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2) testing disparities for small area estimation. Using a Bayesian inseparable space-time model framework, we
estimated the testing positivity rate (TPR) at geographically granular areas of the census block groups (CBGs). Next,
we adopted a rank-based approach to compare the estimated TPR and the testing rate to identify areas with testing deficiency
and quantify the number of needed tests. Applying this to data from Cameron County, Texas, from March 2020 to February 2022,
we identified the CBGs that had experienced substantial testing deficiency, quantified the number of tests that should have been
conducted in these areas, and evaluated the short- and long-term testing disparities. Our proposed analytical framework offers
policymakers and public health practitioners a tool for understanding SARS-CoV-2 testing disparities in geographically small communities.
It could also aid COVID-19 response planning and inform intervention programs to improve goal setting and strategy implementation in
SARS-CoV-2 testing uptake.
(Check our publication:
A novel Bayesian spatial-temporal approach to quantify COVID-19 testing disparities for small area estimation. )
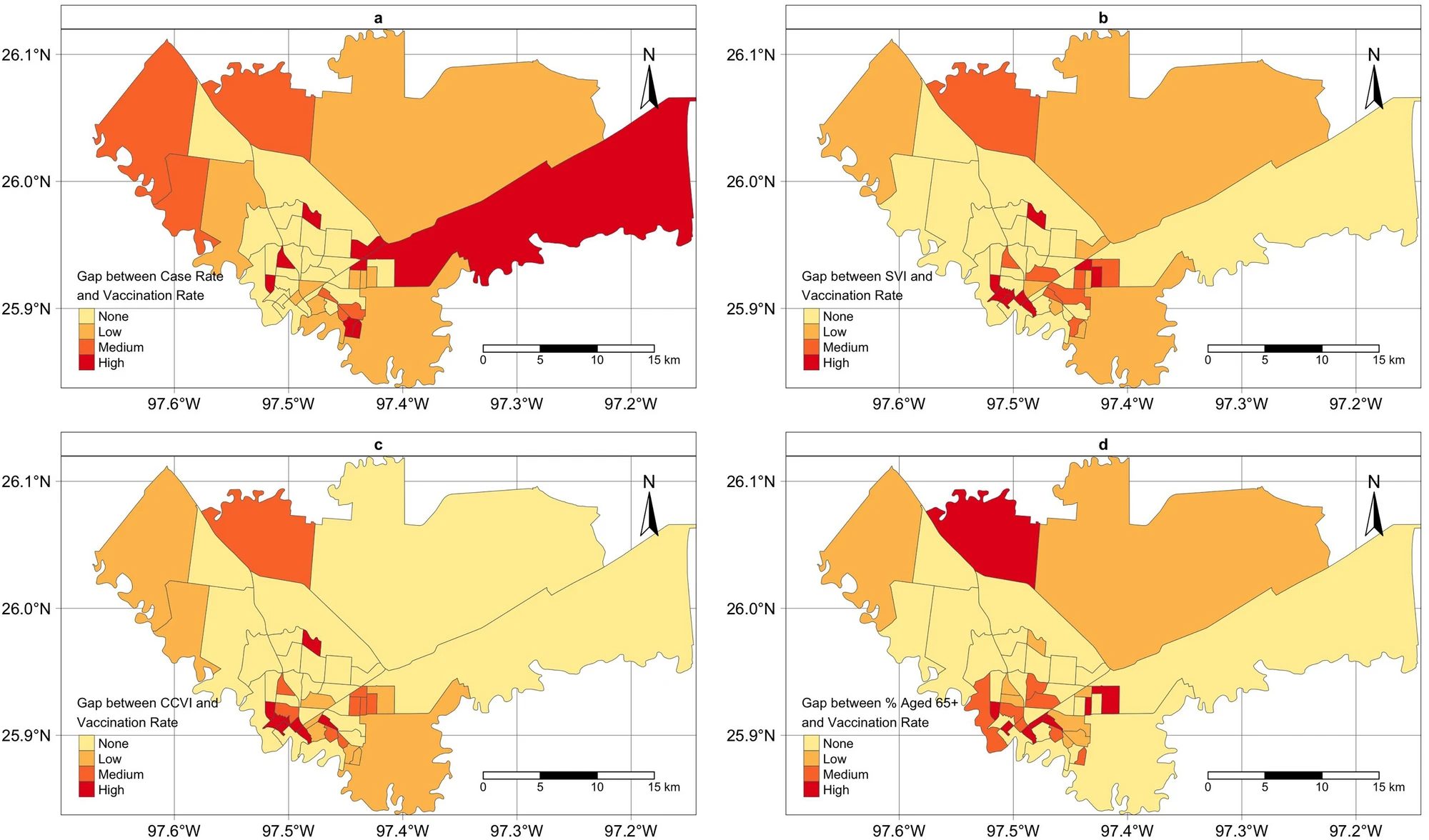
We reported the effective use of real-time geospatial analysis to identify barriers and gaps in COVID-19
vaccination in a minority population living in South Texas on the US-Mexico Border, to inform vaccination
campaign strategies. We developed 4 rank-based approaches to evaluate the vaccination gap at the census tract level,
which considered both population vulnerability and vaccination priority and eligibility. We identified areas with
the highest vaccination gaps using different assessment approaches.
(Check our publication:
Real-time geospatial analysis identifies gaps in COVID-19 vaccination in a minority population. )
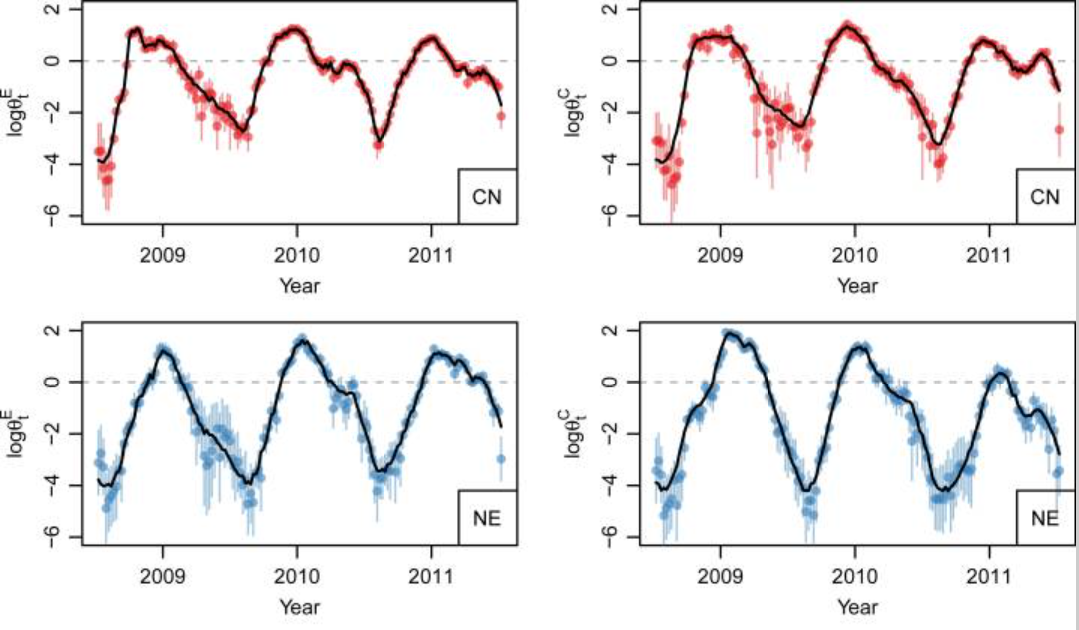
We extend an interesting class of space–time models for infectious disease data proposed by Held and co-workers,
to analyse data on hand, foot and mouth disease, collected in the central north region of China over 2009–2011.
We provide a careful derivation of the model and extend the model class in two directions. First, we model the
disease transmission between age–gender strata, in addition to space and time. Second, we use our model for
inference on effective local reproductive numbers. (Check our publication:
Stratified space–time infectious disease modelling, with an application to hand, foot and mouth disease in China)
We develop a simple method to quickly obtain weekly stratum-specific estimates of the total number of cases attributable to each pathogen,
even when there are few cases subsampled. The estimates can then be used for modeling the pathogen-specific temporal dynamics of interest.
(Check our publication:
Time series modeling of pathogen-specific disease probabilities with incomplete data.)
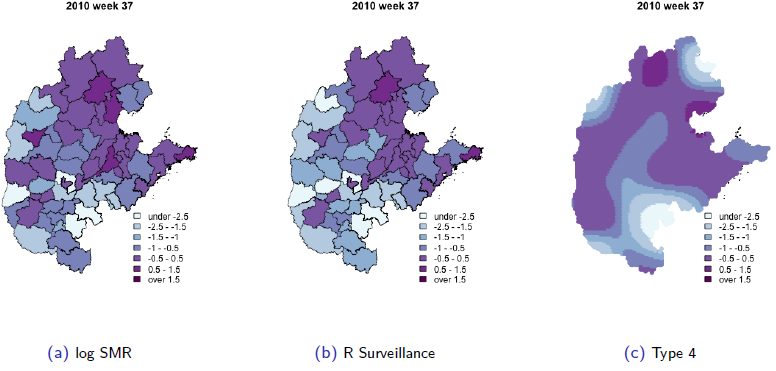
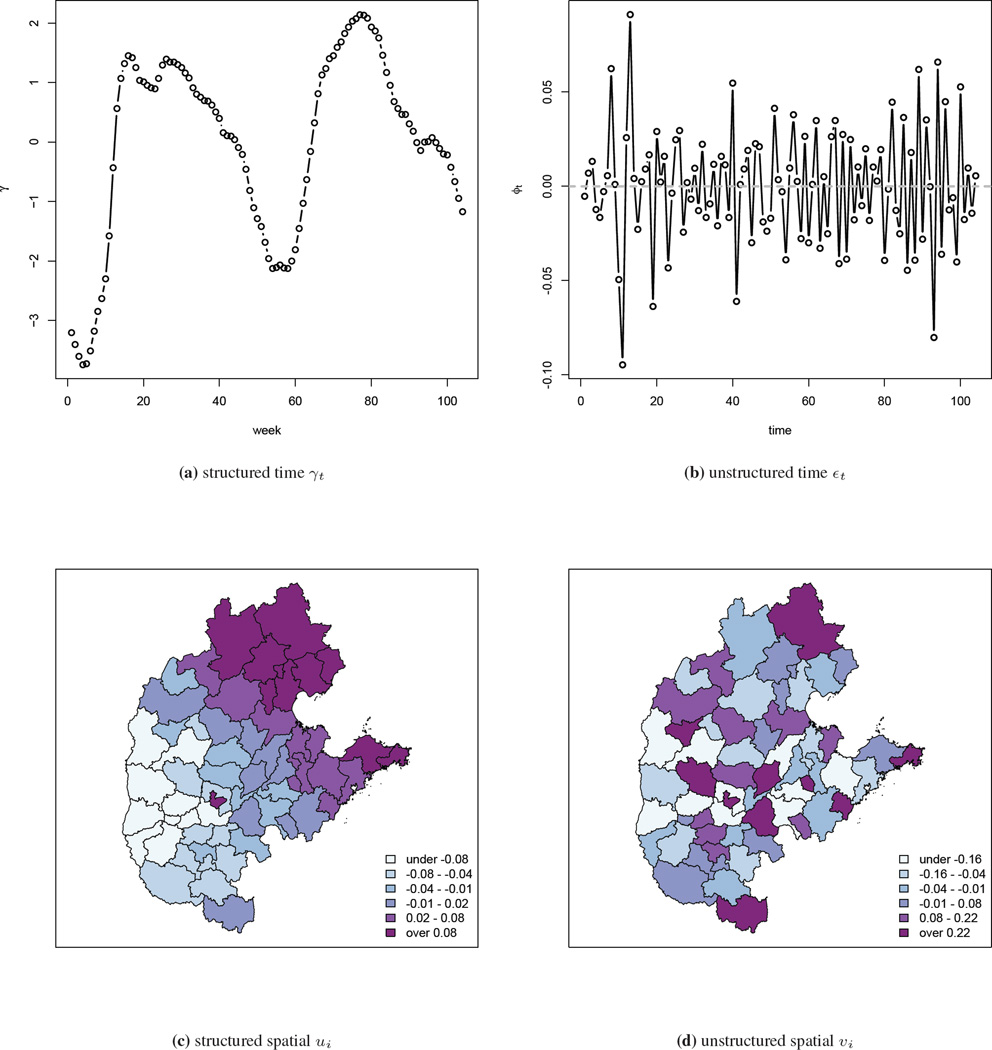
We describe a model that was motivated by the need to analyze hand, foot, and mouth disease surveillance data in China.
The data are aggregated by geographical areas and by week, with the aims of the analysis being to gain insight into the space-time
dynamics and to make short-term predictions, which will aid in the implementation of public health campaigns in those areas with a
large predicted disease burden. The model we develop decomposes disease-risk into marginal spatial and temporal components and a
space-time interaction piece. The latter is the crucial element, and we use a tensor product spline model with a Markov random field
prior on the coefficients of the basis functions.
(Check our publication:
Bayesian penalized spline models for the analysis of spatio-temporal count data.)